The interplay between genetic and environmental factors plays a critical role in the development of type 1 diabetes (T1D). While variants in non-HLA coding regions have been identified as potential therapeutic targets, their exact contribution to the pathophysiology of disease remains unclear. The aim of the study was to characterize six non-HLA variants in pediatric patients with T1D and explore their potential association with clinical parameters and other autoimmune diseases, such as thyroiditis and celiac disease (CD).
MethodsWe analyzed six variants (c.1858T>C, c.49A>G, c.919A>G, c.784T>C, c.461G>A, and c.-17-6T, located in the PTPN22, CTLA4, CD226, SH2B3, FUT2 and INS genes) in a pediatric sample (age ≤ 18 years) with T1D, comparing it with a CD group and a control group. The variants were genotyped using quantitative PCR with TaqMan probes.
ResultsWe observed that variants in PTPN22, CD226 and INS were overrepresented in patients with T1D compared to the control and CD groups. There was a significant association between the presence of anti-glutamate decarboxylase autoantibodies (GADA) and the CTLA4 variant (P = .005), as well as between insulinoma-associated anti-tyrosine phosphatase autoantibodies (IA2A) and the PTPN22 variant (P < .03).The number of positive pancreatic autoantibodies was associated with the FUT2 variant (P = .02). Additionally, age at onset was associated with CTLA4 (P = .01) and SH2B3 (P < .05) variants.
ConclusionThe analyzed variants in the PTPN22, CD226, and INS genes were overrepresented in pediatric patients with T1D, suggesting potential therapeutic targets for modulating the autoimmune process. Their associations with specific clinical and autoimmune profiles can be applied in the identification of high-risk patients and help optimize their follow-up.
La interacción entre factores genéticos y ambientales es clave en el desarrollo de la diabetes tipo 1 (DM1). Aunque las variantes en la región codificante no-HLA son potenciales marcadores para terapias dirigidas, su contribución a la enfermedad es poco conocida. Nuestro objetivo fue caracterizar 6 variantes no-HLA en pacientes pediátricos con DM1, analizando su asociación con parámetros clínicos y otras patologías autoinmunes (tiroiditis y enfermedad celiaca –EC–).
Pacientes y métodosSe analizaron las variantes c.1858T>C, c.49A>G, c.919A>G, c.784T>C, c.461G>A y c.-17-6T>A en los genes PTPN22, CTLA4, CD226, SH2B3, FUT2 e INS en una población pediátrica con DM1 (≤18a), comparándola con un grupo control y otro con EC. Las variantes se identificaron mediante PCR cuantitativa usando sondas Taqman.
ResultadosObservamos sobrerrepresentación de las variantes enPTPN22, CD226 e INS en los pacientes con DM1 frente a los grupos con EC y control. Encontramos asociación entre la presencia de autoanticuerpos antiglutamato descarboxilasa (GADA) y la variante en CTLA4 (p=0,005), y entre autoanticuerpos antitirosina fosfatasa asociados a insulinoma (IA2A) y la variante en PTPN22 (p<0,03). El número de autoanticuerpos pancreáticos positivos mostró asociación con la variante en FUT2 (p=0,02). La edad al debut se asoció con las variantes en CTLA4 (p=0,01) y en SH2B3 (p<0,05).
ConclusionesLos pacientes pediátricos con DM1 tienen sobrerrepresentadas las variantes analizadas en los genes PTPN22, CD226 e INS, señalando posibles dianas terapéuticas que modulen el proceso autoinmune. Su asociación con determinados perfiles clínicos y autoinmunes podría ayudar a identificar pacientes de riesgo y optimizar su seguimiento.
Type 1 diabetes (T1D) is the most common chronic metabolic disorder in children and is characterized by hyperglycemia secondary to insulin deficiency resulting from the immune-mediated destruction of pancreatic ß cells.1 Based on data published by the International Diabetes Federation, in 2023 Spain was one of the ten countries with the highest prevalence of T1D in every age group, with 17 245 inhabitants aged less than 20 years with T1D.2 The current data indicate an annual increasing trend of 2.5%,3 and this is one of the diseases with the highest prevalence and greatest impact on the pediatric population. A significant increase in the frequency of T1D is expected by 2040, especially in developing countries.4
The main biomarkers for T1D are the autoantibodies to glutamate decarboxylase (GADA), protein tyrosine phosphatase-like insulinoma-associated antigen-2 (IA2A) and insulin (IAA).5 More recently, autoantibodies against zinc transporter 8 (ZnT8) have been described and become widely used as an additional marker in the characterization of type 1 diabetes.6 In addition, T1D is frequently associated with other autoimmune diseases, such as thyroiditis and celiac disease (CD).7
Type 1 diabetes is a polygenic disease in which the interaction between genetic and environmental factors plays a crucial role in the development of the autoimmune process. Over half of the total genetic risk has been attributed to variants involving the human leukocyte antigen (HLA) class II gene region and the insulin (INS) gene locus, which are associated with dysregulation of T cell-mediated autoimmunity,7 while the other half is attributed to non-HLA variants.8 This distribution evinces the importance of studying non-HLA variants to understand the genetic predisposition to T1D.
In recent years, several genome-wide association studies (GWAS) have led to the identification of multiple non-HLA genetic risk variants that seem to play a role in the clinical heterogeneity of T1D.9,10 More than 90 risk alleles associated with the disease have been identified to date.11 Although many of these risk alleles are located in intronic—and therefore, noncoding—regions,9 a small percentage are located in coding regions and could be potential markers for targeted therapies. A recent review by Shapiro et al.10 analyzed 13 coding single nucleotide polymorphisms (SNPs) in 10 different genes (PTPN22, IFIH1, SH2B3, CD226, TYK2, FUT2, SIRPG, CTLA4, CTSH and UBASH3A), describing not only the association of these variants with the risk of T1D, but also their potential clinical impact and therapeutic interest in the context of precision medicine. These variants affect the progression of pediatric autoimmunity at different stages: for example, the CTLA4 variant is associated with rapid progression of autoimmunity,11 while the SH2B3 variant may be associated with the initial development of autoantibodies.12 Furthermore, many of these T1D-associated variants have been linked to other autoimmune diseases, such as thyroiditis and celiac disease, which is consistent with the frequent occurrence of polyautoimmunity in the pediatric population.13 The co-occurrence of autoimmune diseases has clinical and translational relevance, as therapies targeting these variants could be beneficial in multiple diseases.
The recent development of novel therapies capable of slowing, at least temporarily, the progression to clinical stages of the disease underscores the need for early and accurate diagnosis. One approach to translating genetic data into a predictive measure of disease susceptibility is to add up the risk effects of both HLA variants and non-HLA polymorphisms into a polygenic risk score.
In this context, we decided to characterize five risk alleles (rs2476601, rs231775, rs763361, rs3184504 and rs601338) located in coding regions of non-HLA genes PTPN22, CTLA4, CD226, SH2B3 and FUT2 and an intronic variant of the INS gene in a cohort of pediatric patients with T1D. The objectives of our study were to estimate the prevalence of the genetic variants analyzed in the sample and compare it with a control population, in addition to analyzing their potential association with clinical variables (age at onset, T1D autoantibodies at onset), and other autoimmune diseases (thyroiditis and CD). This would allow us to investigate the potential relationship between these variants and the heterogeneous clinical presentation of these patients and to identify potential markers for targeted therapies that could improve the quality of life of patients with T1D within a personalized medicine approach.
Patients and methodsWe conducted a retrospective, cross-sectional, observational and analytical study based on the analysis of non-HLA variants associated with T1D risk in a cohort of pediatric patients (age ≤ 18 years) with T1D diagnosis followed up at a tertiary care hospital. Since the genetic variants under study have been described in other autoimmune diseases, such as CD, we also analyzed the prevalence of these variants in a series of patients with CD without T1D managed in the pediatric gastroenterology department of the same hospital. In addition, we included a control group of pediatric patients managed in our hospital without autoimmune diseases, whose allele frequencies were similar to those reported for the Caucasian population in the Database of Single Nucleotide Polymorphisms (dbSNP) of the National Institutes of Health of the United States.
We recruited patients with T1D the Pediatric Endocrinology and Diabetes Clinic of a tertiary care hospital and obtained signed informed consent in accordance with current regulations. The study was approved by the Ethics Committee of the hospital. We excluded patients for whom we did not obtain informed consent, whose collected samples were inadequate for the study or in whom venipuncture could not be performed, patients with chromosomal abnormalities (Down syndrome, Turner syndrome, etc.) and patients with familial polyendocrinopathies. In the CD group and the control group, we used the leftover specimens from routine follow-up testing for the study tests.
The clinical data of the patients were retrieved from the health records and entered into a database created for the study. We collected data on the following variables: age at onset of T1D, number of positive autoantibodies (GADA, IA2A, IAA), presence or absence of antithyroid antibodies (thyroid peroxidase [TPO] and/or thyroglobulin [TGA] antibodies), and presence or absence of CD and/or hypothyroidism.
Pancreatic antibodies were measured by enzyme-linked immunosorbent assay (ELISA) on the Triturus analyzer (Grifols), and antithyroid antibodies were measured by chemiluminescent immunoassay on the Atellica Solution analyzer (Siemens Healthineers, Munich, Germany).
The specimens used for testing were samples of anticoagulated whole blood collected in K3-EDTA tubes. DNA was extracted with the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) for subsequent quantification in a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Last of all, DNA samples were stored in 2mL cryotubes at −80°C until use.
Variant identification was carried out with 5 predesigned TaqMan assays for variants in genes PTPN22 (C__16021387_20), CTLA4 (C___2415786_20), CD226 (C___1464836_20), SH2B3 (C___2981072_10), INS (C___1223317_10) and one custom-designed assay (ThermoFisher Scientific) for a FUT2 variant (ANWDEJA) using a Step One Plus quantitative PCR system (Applied Biosystems, Foster City, CA, USA) (Table 1).
Non-HLA risk variants analyzed in the study.
| Assay | Gene | rsID | Change |
|---|---|---|---|
| C__16021387_20 | PTPN22 | rs2476601 | p.Trp620Arg |
| c.1858T>C | |||
| C___2415786_20 | CTLA4 | rs231775 | p.Thr17Ala |
| c.49A>G | |||
| C___1464836_20 | CD226 | rs763361 | p.Ser307Gly |
| c.919A>G | |||
| C___2981072_10 | SH2B3 | rs3184504 | p.Trp262Arg |
| c.784T>C | |||
| ANWDEJA | FUT2 | rs601338 | p.Trp154Ter |
| c.461G>A | |||
| C___1223317_10 | INS | rs689 | c.-17-6T>A |
Abbreviations: CD226, cluster of differentiation 226 gene; CTLA4, cytotoxic T-lymphocyte associated protein 4 gene; FUT2, gene encoding α(1,2)-fucosyltransferase 2; INS, insulin gene; PTPN22, protein tyrosine phosphatase non-receptor type 22 gene; SH2B3, SH2B adapter protein 3 gene.
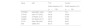
Association between different variants and the clinical and autoimmune profile in the group of patients with type 1 diabetes without celiac disease.
| Gene | rsID | Age of onset | Autoimmunity (GADA/IA2A/AIA) | Abs at onset | Celiac disease (n=16) | TG and/or TPO Abs (n=37) |
|---|---|---|---|---|---|---|
| Number of Abs at onset (n=173) | ||||||
| PTPN22 | rs2476601 (G/A) | NS | IA2A (P=.026) | NS | NS | NS |
| CTLA4 | rs231775 (A/G) | P=.010 | GADA (P=.005) | NS | NS | NS |
| CD226 | rs763361 (C/T) | NS | NS | NS | P=.084 | NS |
| SH2B3 | rs3184504 (C/T) | P=.047 | NS | NS | NS | P=.072 |
| FUT2 | rs601338 (G/A) | NS | NS | P=.017 | NS | NS |
| INS | rs689 (A/T) | NS | NS | NS | NS | NS |
Abbreviations: Abs, autoantibodies; CD226, cluster of differentiation 226 gene; CTLA4, cytotoxic T-lymphocyte associated protein 4 gene; FUT2, gene encoding α(1,2)-fucosyltransferase 2; GADA, glutamate decarboxylase autoantibodies; IAA, anti-insulin autoantibodies; IA2A, insulinoma-associated tyrosine phosphatase autoantibodies; INS, insulin gene; NS, not significant; PTPN22, protein tyrosine phosphatase non-receptor type 22 gene; SH2B3, SH2B adapter protein 3 gene; TG, thyroglobulin; TPO, thyroid peroxidase.
We compared allele frequencies with both a control group in our hospital (n=50; age ≤ 18 years) and the population-level allele frequency in the dbSNP for the European population, as this was the ancestry of 86.7% of the probands in our sample.
In the descriptive analysis, we expressed quantitative variables as median (IQR). In the comparative analysis, we used the χ2 test to assess the association between each of the non-HLA variants and clinical variables, the nonparametric Kruskal-Wallis test to establish the distribution of the age of onset in relation to each variant, and the Mann-Whitney U test to compare two independent groups. For the comparison of allele frequencies, we used the χ2 test or, in the case of very low frequencies, the Fisher exact test. The statistical analysis was performed with the Stata software package, version 15.1 (StataCorp2017, StataCorp, College Station, TX, USA). We considered results with a P value of less than 0.05 statistically significant.
ResultsIn the sample of 194 pediatric patients (age ≤ 18 years) with T1D, 94 were female and 99 were male. The median age at onset was 8.3 years (IQR, 4.5–10.9), with no significant differences by sex. Of the 189 patients for whom pancreatic autoantibody test results were available (GADA, IA2A, and IAA), 168 tested positive for at least one autoantibody (88.83%) and 21 tested negative for all three autoantibodies. Of the 186 patients with thyroid antibody test results, 37 tested positive (TPO or TG antibodies) and nine of them developed hypothyroidism. In addition, 16 of the 194 patients with T1D had a diagnosis of CD (8.2%), and 5 had both thyroid antibodies and CD. It should be noted that autoantibody testing was performed at the time of diagnosis of T1D and that the autoimmune diseases that we report in these patients were clinically confirmed.
Table 3 presents the allele frequencies in the T1D group and the control group. We found overrepresentation of PTPN22, CD226 and INS variants in patients with T1D, with statistically significant differences for CD226 (P=.041) and INS (P=.019) variants.
Frequency of risk alleles in the group of patients with T1D and in the control group.
| Gene | rsID | T1D | Control |
|---|---|---|---|
| Allele frequency (%) | Allele frequency (%) | ||
| (n=178) | (n=50) | ||
| PTPN22 | rs2476601 (G/A) | 12.57 | 7.01 |
| CTLA4 | rs231775 (A/G) | 34.29 | 36.38 |
| CD226 | rs763361 (C/T) | 50.00 | 47.94 |
| SH2B3 | rs3184504 (C/T) | 48.57 | 50.04 |
| FUT2 | rs601338 (G/A) | 48.00 | 48.07 |
| INS | rs689 (A/T) | 83.14 | 72.97 |
Abbreviations: CD226, cluster of differentiation 226 gene; CTLA4, cytotoxic T-lymphocyte associated protein 4 gene; FUT2, gene encoding α(1,2)-fucosyltransferase 2; INS, insulin gene; PTPN22, protein tyrosine phosphatase non-receptor type 22 gene; SH2B3, SH2B adapter protein 3 gene; T1D, type 1 diabetes.
Table 4 presents the odds ratios (ORs) for the different gene variants, reflecting the overrepresentation of some variants in the subset of patients with T1D without CD compared to the subset with CD. These ORs were not adjusted for age, sex or HLA genotype.
Risk variants analyzed in the study with their respective odds ratios in patients with type 1 diabetes without celiac disease and patients with celiac disease without type 1 diabetes.
| Gene | rsID | OR T1D without CD | OR CD without T1D |
|---|---|---|---|
| PTPN22 | rs2476601 | 2.20 | 1.27 |
| CTLA4 | rs231775 | 1.03 | 0.77 |
| CD226 | rs763361 | 1.17 | 1.17 |
| SH2B3 | rs3184504 | 1.28 | 0.85 |
| FUT2 | rs601338 | 0.86 | 0.67 |
| INS | rs689 | 2.21 | 1.16 |
Abbreviations: CD, celiac disease; CD226, cluster of differentiation 226 gene; CTLA4, cytotoxic T-lymphocyte associated protein 4 gene; FUT2, gene encoding α(1,2)-fucosyltransferase 2; INS, insulin gene; OR, odds ratio; PTPN22, protein tyrosine phosphatase non-receptor type 22 gene; SH2B3, SH2B adapter protein 3 gene; T1D, type 1 diabetes.
Table 2 presents the results for the association between different gene variants and clinical features. We found an association between the presence of GADA and the CTLA4 variant (P=.01), and between the presence of IA2A and the PTPN22 variant (P<.03). The age of onset was associated with the CTLA4 (P=.010) and SH2B3 (P<.05) variants. There was also an association between the FUT2 variant (P=.017) and a greater number of positive pancreatic autoantibodies at onset. We did not find an association between the CD226 variant and CD (P=.084) nor between the SH2B3 variant and the presence of thyroid antibodies (P=.072).
In addition to the 194 pediatric patients with T1D (16 with both T1D and CD), we recruited 115 patients with CD without T1D (63 female, 52 male). The median age at diagnosis was 11.3 years (IQR, 7.57–14.4). All patients in the study were classified into four categories: T1D without CD, T1D with CD, CD without T1D, and control group. Fig. 1 compares the allele frequencies of the variants under study in these four groups. The variants in PTPN22, CD26, and INS were overrepresented in patients with T1D compared to the other groups. The statistical analysis revealed significant differences in the CD226 (P=.041) and INS (P=.019) variants between the group of patients with T1D without CD and the control group. In addition, when we compared the group of patients with T1D and CD to the control group, we found significant differences in the CD226 variants (P=.008). On the other hand, we did not find statistically significant differences between the group of patients with CD without T1D and the control group nor between the T1D without CD and the T1D with CD groups.
DiscussionThe results obtained are consistent with the findings of other studies, including large-scale studies, such as TEDDY and TrialNet,10,14–18 that describe non-HLA variants associated with an increased risk of T1D, as we found that some of them (in INS, PTPN22 and CD226 loci) were overrepresented in our group of patients with T1D. We found significantly differences in the allele frequency of some of these variants between groups (T1D without CD, T1D with CD, CD without T1D and control) (Fig. 1). It should be noted that the frequency of CD and T1D detected in our study (8.2%) was similar to those described by other authors.19 These findings corroborate the polygenic nature of the disease and point to potential therapeutic targets that modulate the autoimmune process. On the other hand, the overrepresentation of some variants, such as the CD226 variant found in patients with more than one autoimmune disease (T1D and CD), could indicate a distinctive susceptibility profile in a subset of patients.
Thus, the most frequent variant at the PTPN22 locus was rs2476601, in which in which adenine (A) substitutes for the wild-type guanine (G) and gives rise to the change p.Arg620Trp. This variant is considered the most significant risk factor for T1D following variants in the HLA-DR/DQ and the INS loci, with an OR of 1.81.10 In our study, this variant was overrepresented in patients with T1D compared to both the control group and the group of patients with CD without T1D. According to GWAS results, the presence of this variant appears to be associated with the presence of IAA in the early stages of disease,15 as well as the development of other autoimmune diseases and early onset of T1D.16,17 Although we did not observe this association in our sample, we did find an association between this variant and the presence of IA2A, described in stages in which pancreatic beta cell destruction is already present, which had not been reported before. These differences could be due to patients in our cohort being at a different stage of disease compared to other studies or to differences between the study populations.
Similarly, the variant rs231775 in CTLA4, in which the substitution of A for G results in the amino acid change p.Thr17Ala, seems to be associated with the presence of GADA and late disease onset.20,21 Our study confirmed the association between the presence of GADA and older age at onset in patients with this variant. The involvement of this CTLA-4 receptor in T cell regulation has sparked interest in the use of the drug Abatacept, a CTLA4-IgG1 fusion protein that blocks T cell activation, to delay the loss of beta cell function in newly diagnosed T1D patients.22 Thus, several clinical trials are currently underway to evaluate the usefulness of this molecule, either alone or in combination with Rituximab (anti-CD20), in preventing or delaying the progression of T1D in its early stages.23
The rs3184504 variant in SH2B3 entails a substitution of C for T that results in the amino acid change R262W. In other studies, this variant has been mainly associated with the development of autoantibodies against pancreatic beta cells.12 In our study, we found an association between this variant and a later age of onset of T1D. In addition, we found that this variant was less frequent in patients with CD (with or without T1D). This could be related to its characterization as a potential protective factor against CD, reducing the risk of developing this disease.11
Lastly, when it came to variant rs601338 in FUT2, in which adenine is substituted for guanine giving rise to the change p.W154*, we found a statistically significant association with the presence of at least one autoantibody at onset of T1D. The detection of this variant and its association with autoantibodies at onset is relevant, as previous studies have found a steeper decline in the first-phase insulin response during the glucose tolerance test in children with multiple autoantibodies and carrying this particular variant.24 Furthermore, according to some authors, its clinical relevance lies in the fact that its presence in patients with T1D increases susceptibility to changes in the gut microbiota and inflammatory status, conditions that can be mitigated with early interventions such as adapted diets, prebiotics, probiotics, or formula enriched with α1,2-fucosyloligosaccharides.25
The main limitation of the study is the limited number of patients in the different subgroups, such as those with T1D and CD or with T1D and hypothyroidism, due to the real-world prevalence of these conditions in the pediatric population. Likewise, the genetic analysis included a limited set of variants and did not cover all the variants described in the previous literature. Another limitation is that we did not analyze the association of these variants with the presence of ZnT8 autoantibodies. Another possible limitation is that some low-prevalence variants may not be identified in a sufficient number of patients to achieve statistically significant results. To address this challenge, our group continues to increase the sample size. In addition, we emphasize the need to conduct multicenter studies with greater statistical power to conclusively confirm our results.
It is worth noting that identifying the genes where risk variants occur can help tailor the most appropriate therapy to each patient's profile, including risk estimation formulas.26–28 Thus, patients who carry risk variants in PTPN22, CD226 and CTLA4 could benefit from drugs modulating T-cell costimulation.10 Similarly, there is interest in therapies for induction of antigen-specific tolerance in patients with risk variants in PTPN22 and CTLA4, due to their propensity to develop IAA and GADA, respectively, in the early stages of T1D.15
Although the identified non-HLA variants are promising as biomarkers, their individual clinical utility remains limited and requires validation in independent, multicenter cohorts.
ConclusionsThe observed overrepresentation in pediatric patients with T1D of some of the analyzed risk variants for T1D in the PTPN22, CD226 and INS genes support the polygenic nature of the disease and suggest potential therapeutic targets that may modulate the autoimmune process.
The observed association between certain non-HLA risk variants and specific clinical and autoimmune profiles, as well as the age of T1D onset, could help identify at-risk patients and guide follow-up and management.
The overrepresentation of some variants (in CD226) in patients with more than one autoimmune disease (T1D and CD) could indicate a distinctive susceptibility profile in a subset of patients.
Future studies should include larger samples and analyze additional non-HLA genetic variants associated with T1D and other autoimmune diseases. It would also be interesting to study the allelic and autoimmune profiles of siblings of patients with T1D, with or without CD, and to analyze HLA class II risk factors in T1D in order to calculate risk scores combining HLA and non-HLA variants and other previously identified clinical and risk profiles. In addition, comparing the variants identified in the minority of patients in whom autoantibodies are not detected with those identified in patients in whom they are detected could contribute information on differences that may explain the progression of the disease.
FundingThis study was partially funded with the José Igea 2023 grant.
The authors have no conflicts of interest to declare.